Alvaro Mallagaray's research group
Our research focuses on the development and application of NMR-based methods for the molecular characterization of biological systems. Central themes include metabolic and glycosylation-related signatures in biological fluids and their alterations in inflammatory, oncogenic, and infectious diseases, with the goal of biomarker discovery and validation. A further focus lies on structural glycobiology and virology. We investigate the molecular mechanisms of glycan recognition by viruses such as noroviruses and coronaviruses, as well as the processing of complex polysaccharides and N-glycans in the human gut microbiome. By combining experimental NMR spectroscopy with computational data analysis and interdisciplinary collaborations, we pursue a translational approach spanning basic research to clinical applications.
Previous and current research
Project 1: Fast quantification of plasma N-glycosylation profiles associated to cancer progression and inflammatory processes by NMR
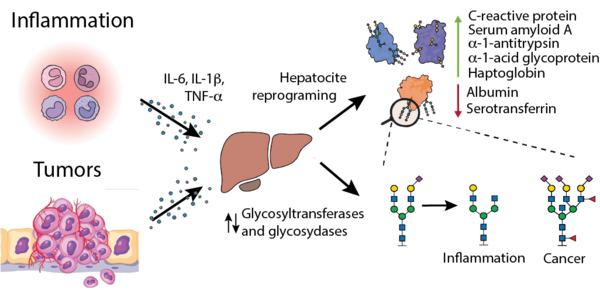
Protein glycosylation is the most common posttranslational modification in human cells, resulting in a diverse array of glycoconjugates. Aberrant glycosylations in circulating proteins are used as potent diagnostic parameters for cancer due to neoplasia-induced hepatic reprogramming, and have been observed in many inflammation-related diseases.
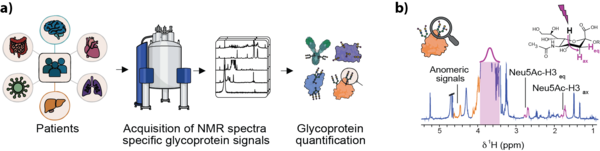
Within this project we develop rapid, quantitative NMR methods to characterize inflammation-related glycoprotein profiles directly in native human serum/plasma, with the goal of translating glycosylation-informed biomarkers into scalable clinical diagnostics and large cohort studies. We focus on extracting protein-specific and glycosylation-specific information from standard 1D NMR spectra with minimal sample handling, while keeping the workflow robust, automated, and reproducible across instruments and sites.
In addition to signals from small metabolites and lipoproteins, blood NMR spectra also contain characteristic resonances from protein-bound glycans, as it is the case of GlycA/GlycB. In addition to them, NMR spectra of blood contain a plethora of glycan-derived signals informing about glycan composition, and responding strongly to inflammatory and oncogenic processes. Our work focuses on creating new experiments and informatic tools aiming to the extraction of this hitherto unexplored NMR signals, allowing novel mechanistic interpretations related to changes in glycoprotein concentrations, as well as glycoprofiles suitable for biomedical studies.
The development of new biomarkers is driven by our close collaboration with clinical partners and the systematic translation of our work into diverse cohort studies. Using our patented NMR-based technology for assigning and quantifying glycoprotein-specific contributions and glycosylation patterns in blood serum and plasma, we aim to identify and validate biomarker profiles across a broad spectrum of inflammatory and oncogenic pathologies. These include, among others, inflammatory bowel disease, Parkinson’s disease, systemic sclerosis, MASLD and its progression to hepatocellular carcinoma, as well as additional cancers such as colorectal carcinoma, cholangiocarcinoma, uveal melanoma, and multiple myeloma.
Project 2: Control of complex polysaccharide metabolism in the human gut microbiome
The gut microbiome plays a central role in human health by supporting digestion, shaping the immune system, and helping to protect against pathogens. Many diseases have been linked to imbalances in these microbial communities ('dysbiosis'). A major factor determining which microbes thrive in the intestine is the constant supply of complex carbohydrates from our diet and from the mucus layer that lines the gut. Because these sugars are a key energy source, gut bacteria have evolved highly specialized ways to break down, capture, and import them, yet many of the underlying molecular mechanisms are still not well understood.
This project aims to reveal, at a molecular level, how gut bacteria process complex polysaccharides and N-glycans. Certain gut bacteria (especially Bacteroidetes) organize the required genes in compact 'toolboxes' called polysaccharide utilization loci (PULs), which encode enzymes that cut specific carbohydrates and proteins that help bind and transport the resulting fragments into the cell. Understanding these pathways is important because it can open new therapeutic strategies to steer the microbiome—for example, supporting beneficial bacteria, limiting harmful ones, and reducing the growth of inflammatory or antibiotic-resistant strains.
We focus on NMR combined with complementary structural biology techniques (mass spectrometry, cryo-EM, crystallography, etc.) to determine how key enzymes work, how they recognize their carbohydrate targets, and how different proteins cooperate to move sugars from the gut environment into bacterial cells. By mapping these mechanisms in detail, we aim to build a foundation for designing precision interventions that reshape microbial metabolism—and ultimately microbiome composition—in a controlled and medically useful way.
Project 3: Study of the interaction of Norovirus with cell attachment factors required for infection
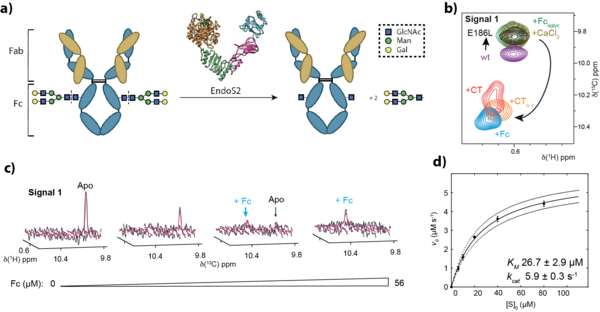
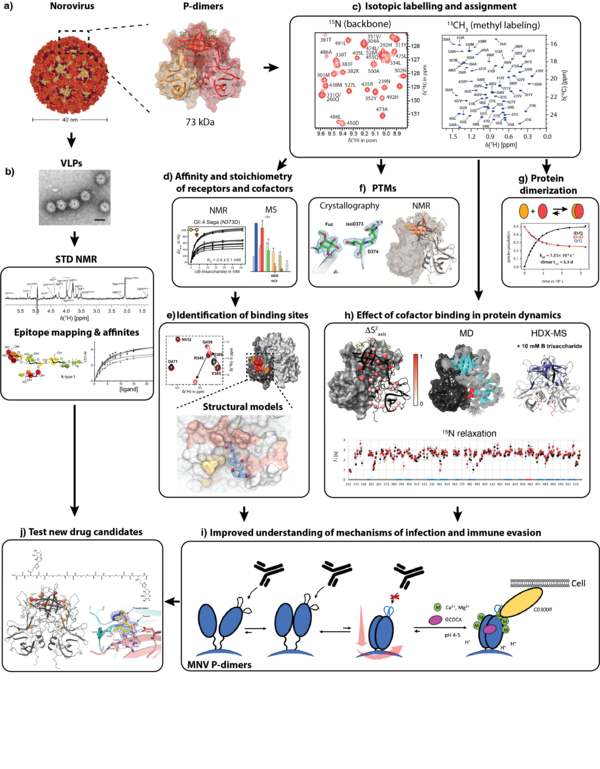
Noroviruses are a leading cause of acute gastroenteritis, and infection begins with capsid attachment to histo-blood group antigens (HBGAs) on the gastrointestinal mucosa. In this integrative project we use NMR-driven structural glycobiology to define how human and murine norovirus capsid proteins recognize host carbohydrates and bile-acid cofactors required for infection in order to develop highly selective entry inhibitors against Norovirus.
Our core strategy combines ligand-observed and protein-observed NMR. Saturation transfer difference NMR titrations provide ligand epitope mapping and binding readouts in both P-dimers and assembled virus-like particles (VLPs), while 1H,15N-TROSY-HSQC chemical-shift perturbation (CSP) titrations on uniformly 2H,15N-labeled P-domains serve as a stringent, direct proof of binding under near-physiological conditions. To access larger systems and subtle allosteric effects, we implemented methyl-TROSY experiments on selectively methyl-labeled capsid domains and integrated orthogonal validation by mass spectrometry, X-ray crystallography and long-timescale molecular dynamics in collaboration with expert groups.
Across multiple human and murine norovirus strains, we established that prevalent GII.4 capsid domains bind HBGAs independently with low-mM affinities, requiring fucose as the minimal motif and showing no recognition of non-fucosylated sialoglycans; we also uncovered key sources of misleading 'complex binding', including glycan clustering in native mass spectroscopy and a time-dependent N373→isoAsp conversion that can weaken HBGA binding. Extending beyond glycans, combined backbone and 13C methyl-labelling NMR with molecular dynamics revealed a low-affinity, flexible bile-acid pocket in GII.4 capsids. Overall, this project redefined key aspects of norovirus ligand recognition, provided best-practice guidelines for reproducible binding studies across platforms, and delivered mechanistic foundations for rational entry-inhibitor concepts, including multivalent glycomaterials and probe-based screening enabled by paramagnetic glycans.