Ulrich Günther's research group
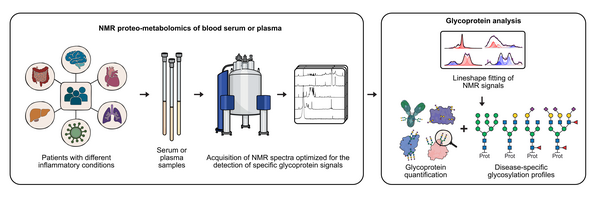
Our research focuses on Nuclear Magnetic Resonance (NMR) spectroscopy and NMR-based metabolomics to investigate metabolic pathways, metabolic fluxes, glycoprotein signatures, and biomarker profiles across a wide range of biological and clinical sample types. We apply and further develop NMR methods for metabolomics and metabolic flux analysis, including screening approaches and the study of metabolic fluxes in cancer cells. Our work also includes the development of new glycoprotein methods, the identification of biomarkers in different clinical cohorts such as cancer, inflammatory bowel disease, NASH, and Parkinson’s disease, and the creation of computational tools for proteo-metabolomics. These developments encompass spectral processing using Matlab, lineshape fitting for quantitative analysis, and statistical evaluation of NMR data. A further part of our research is dedicated to deciphering NMR signatures of different sample materials, ranging from blood serum and plasma to urine and stool.
Previous and current research
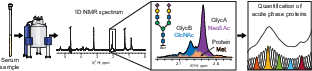
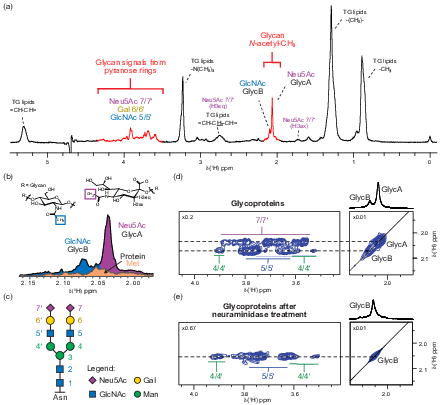
NMR spectroscopy is a well-established and powerful method for metabolomics of biological samples, particularly blood. Blood NMR spectra contain signals from small-molecule metabolites, lipoproteins, and two characteristic resonances originating from acetyl groups of protein-bound glycans, known as GlycA and GlycB. These signals arise from glycans decorating well-known and highly abundant acute-phase proteins and have been proposed as diagnostic markers, especially in cardiovascular diseases. Their strong response in inflammatory conditions, as demonstrated during COVID-19, highlighted their diagnostic relevance and motivated us to study the origin and nature of these glycan-related signals in far greater detail.
In recent work, we developed a new NMR method for the quantitative investigation of glycoprotein signatures in blood serum, opening new opportunities for medical and diagnostic applications. In a publication in Angewandte Chemie, we demonstrated how GlycA/B signals can be deconvoluted to obtain quantitative information about glycosylation types and the identity of the underlying glycosylated proteins. This approach has significant potential for biomarker discovery, as the NMR analysis is rapid, cost-effective, and can be carried out directly on native serum or plasma samples. Overall, our research continues to explore the diagnostic and biological relevance of metabolomic and glycoprotein NMR signatures across diverse clinical contexts.
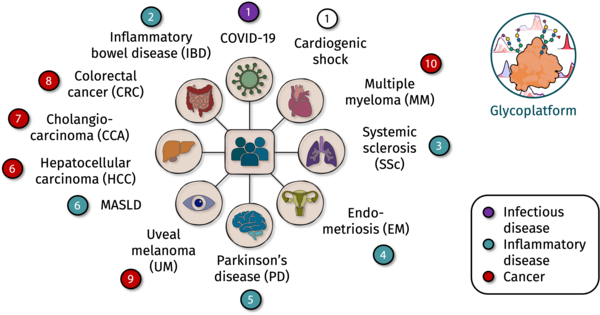
The development of new biomarkers is driven by our close collaboration with clinical partners and the systematic translation of our work into diverse cohort studies. Using our patented NMR-based technology for assigning and quantifying glycoprotein-specific contributions and glycosylation patterns in blood serum and plasma, we aim to identify and validate biomarker profiles across a broad spectrum of inflammatory and oncogenic pathologies. These include, among others, inflammatory bowel disease, Parkinson’s disease, systemic sclerosis, MASLD and its progression to hepatocellular carcinoma, as well as additional cancers such as colorectal carcinoma, cholangiocarcinoma, uveal melanoma, and multiple myeloma.
Our methodological strategy integrates standardized high-throughput measurements (IVDr platform), robust automated signal deconvolution and quantification, and validation against orthogonal reference analytics (e.g., UHPLC–MS). Particularly noteworthy is our close collaboration with Prof. Dr. Astrid Petersmann (Oldenburg) and Prof. Dr. Matthias Nauck (Greifswald): with additional 600-MHz IVDr systems available across our partner institutions, we collectively operate four harmonized high-field platforms. This infrastructure forms the foundation of the North German NMR Alliance, enabling large-scale epidemiological studies to be measured jointly, reproducibly, and at scale—crucial for the identification of robust cross-site biomarkers.